Note
Go to the end to download the full example code.
GeoTIFFs to NetCDF
In this example, we demonstrates the workflow for creating a GS file from the GeoTIFF (.tif/.tiff) file format. This includes adding individual TIF files as single 2-D variables, as well as how to create a 3-D variable by stacking multiple TIF files along a specified dimension.
Additionally, this example shows how to handle Raster data that have differing x-y grids. Specifically, this example creates the following Raster datasets:
- Raster Dataset #1
1a. 2-D magnetic grid, original x-y discretization (600 m cell size)
- Raster Dataset #2
2a. 2-D magnetic grid, aligned to match the x-y dimensions of the resistivity layers (1000 m cell size)
2b. 3-D resistivity grid
Dataset References:
Minsley, B.J., James, S.R., Bedrosian, P.A., Pace, M.D., Hoogenboom, B.E., and Burton, B.L., 2021, Airborne electromagnetic, magnetic, and radiometric survey of the Mississippi Alluvial Plain, November 2019 - March 2020: U.S. Geological Survey data release, https://doi.org/10.5066/P9E44CTQ.
James, S.R., and Minsley, B.J., 2021, Combined results and derivative products of hydrogeologic structure and properties from airborne electromagnetic surveys in the Mississippi Alluvial Plain: U.S. Geological Survey data release, https://doi.org/10.5066/P9382RCI.
import matplotlib.pyplot as plt
from os.path import join
import gspy
from gspy import Survey
from pprint import pprint
Convert data from GeoTIFF to NetCDF
Initialize the Survey
# Path to example files
data_path = "..//data_files//tempest_aseg"
# Survey metadata file
metadata = join(data_path, "data//Tempest_survey_md.yml")
# Establish the Survey
survey = Survey.from_dict(metadata)
container = survey.gs.add_container('derived_products', **dict(content = "raw and processed data",
comment = "This is a test"))
Create the First Raster Dataset Import 2-D magnetic data, discretized on 600 m x 600 m grid Define input metadata file (which contains the TIF filename linked with desired variable name)
d_supp1 = join(data_path, 'data//Tempest_raster_md.yml')
# Read data and format as Raster class object
container.gs.add(key="map", metadata_file=d_supp1)
Create the Second Raster Dataset
# Import both 3-D resistivity and 2-D magnetic data, aligned onto a common 1000 m x 1000 m grid
# Define input metadata file (which contains the TIF filenames linked with desired variable names)
d_supp2 = join(data_path, 'data//Tempest_rasters_md.yml')
# Read data and format as Raster class object
container.gs.add(key="maps", metadata_file=d_supp2)
Save to NetCDF file
d_out = join(data_path, 'tifs.nc')
survey.gs.to_netcdf(d_out)
uuid
title
institution
source
history
references
comment
content
conventions
created_by
type
_FillValue
contractor_project_number
contractor
client
survey_type
survey_area_name
state
country
acquisition_start
acquisition_end
dataset_created
coordinates
_FillValue
time
area
current
frequency
electromagnetic_moment
magnetometer_b_field
electromagnetic_b_field
coordinates
_FillValue
traverse_line_spacing
traverse_line_direction
nominal_terrain_clearance
final_line_kilometers
TRAVERSE LINE NUMBERS
REPEAT LINE NUMBERS
PRE ZERO LINE NUMBERS
POST ZERO LINE NUMBERS
coordinates
_FillValue
aircraft
aircraft_registration
magnetometer
magnetometer_installation
electromagnetic_system
electromagnetic_installation
electromagnetic_coil_orientations
spectrometer_system
spectrometer_installation
radar_altimeter_system
radar_altimeter_sample_rate
laser_altimeter_system
laser_altimeter_sample_rate
navigation_system
navigation_sample_rate
acquisition_system
coordinates
_FillValue
crs_wkt
semi_major_axis
semi_minor_axis
inverse_flattening
reference_ellipsoid_name
longitude_of_prime_meridian
prime_meridian_name
geographic_crs_name
horizontal_datum_name
projected_crs_name
grid_mapping_name
standard_parallel
latitude_of_projection_origin
longitude_of_central_meridian
false_easting
false_northing
authority
wkid
content
comment
type
_FillValue
crs_wkt
semi_major_axis
semi_minor_axis
inverse_flattening
reference_ellipsoid_name
longitude_of_prime_meridian
prime_meridian_name
geographic_crs_name
horizontal_datum_name
projected_crs_name
grid_mapping_name
standard_parallel
latitude_of_projection_origin
longitude_of_central_meridian
false_easting
false_northing
authority
wkid
uuid
comment
content
_FillValue
standard_name
long_name
null_value
valid_range
grid_mapping
coordinates
_FillValue
standard_name
long_name
null_value
valid_range
grid_mapping
coordinates
_FillValue
standard_name
long_name
units
null_value
valid_range
grid_mapping
coordinates
_FillValue
crs_wkt
semi_major_axis
semi_minor_axis
inverse_flattening
reference_ellipsoid_name
longitude_of_prime_meridian
prime_meridian_name
geographic_crs_name
horizontal_datum_name
projected_crs_name
grid_mapping_name
standard_parallel
latitude_of_projection_origin
longitude_of_central_meridian
false_easting
false_northing
authority
wkid
GeoTransform
_FillValue
standard_name
long_name
units
null_value
axis
valid_range
grid_mapping
bounds
standard_name
long_name
units
null_value
valid_range
grid_mapping
_FillValue
standard_name
long_name
units
null_value
axis
valid_range
grid_mapping
bounds
uuid
comment
content
_FillValue
comment
standard_name
long_name
null_value
datum
valid_range
grid_mapping
coordinates
_FillValue
standard_name
long_name
null_value
valid_range
grid_mapping
coordinates
_FillValue
standard_name
long_name
null_value
valid_range
grid_mapping
coordinates
_FillValue
standard_name
long_name
units
null_value
valid_range
grid_mapping
coordinates
_FillValue
standard_name
long_name
units
null_value
valid_range
grid_mapping
coordinates
_FillValue
crs_wkt
semi_major_axis
semi_minor_axis
inverse_flattening
reference_ellipsoid_name
longitude_of_prime_meridian
prime_meridian_name
geographic_crs_name
horizontal_datum_name
projected_crs_name
grid_mapping_name
standard_parallel
latitude_of_projection_origin
longitude_of_central_meridian
false_easting
false_northing
authority
wkid
GeoTransform
_FillValue
comment
standard_name
long_name
units
null_value
axis
positive
datum
valid_range
grid_mapping
bounds
standard_name
long_name
units
null_value
valid_range
grid_mapping
_FillValue
standard_name
long_name
units
null_value
axis
valid_range
grid_mapping
bounds
_FillValue
standard_name
long_name
units
null_value
axis
valid_range
grid_mapping
bounds
Reading back in the GS NetCDF file
new_survey = gspy.open_datatree(d_out)['survey']
Plotting
# Make a map-view plot of a specific data variable, using Xarray's plotter
# In this case, we slice the 3-D resistivity variable along the depth dimension
new_survey['derived_products']["maps"]['resistivity'].plot(col='z', vmax=3, cmap='jet', robust=True)
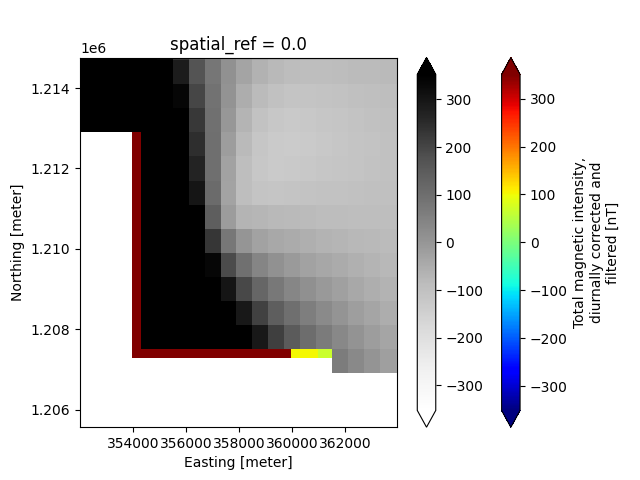
# Make a map-view plot comparing the different x-y discretization of the two magnetic variables, using Xarray's plotter
plt.figure()
ax=plt.gca()
new_survey['derived_products']["maps"]['magnetic_tmi'].plot(ax=ax, cmap='jet', robust=True)
new_survey['derived_products']["map"]['magnetic_tmi'].plot(ax=ax, cmap='Greys', cbar_kwargs={'label': ''}, robust=True)
plt.ylim([1.20556e6, 1.21476e6])
plt.xlim([3.5201e5, 3.6396e5])
plt.show()
Total running time of the script: (0 minutes 0.568 seconds)